Biophysics: A tug-of-war with corona
1 Apr 2022
How stable are the bonds formed by coronaviruses with human cells? Researchers from LMU have designed a new assay to find out – and to investigate drugs made to prevent the virus from binding.
1 Apr 2022
How stable are the bonds formed by coronaviruses with human cells? Researchers from LMU have designed a new assay to find out – and to investigate drugs made to prevent the virus from binding.
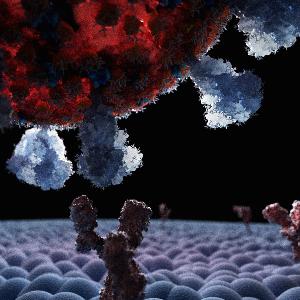
SARS-CoV-2 virus binding to a cell surface | © Chris Hohmann/LMU
Most coronavirus infections begin in the mouth and throat, where SARS coronaviruses bind to the human ACE2 receptor before introducing their genetic material into cells. After initial binding to the receptor, however, the viruses are exposed to numerous forces: breathing, coughing, and sneezing, for example, generate air currents through the mouth and nose and can dislodge the viruses from potential host cells. From the perspective of the virus, it is therefore essential that its binding to ACE2 can withstand external forces to enable successful infections.
The groups led by LMU Professors Hermann Gaub and Jan Lipfert are experts in “force spectroscopy,” a collective term for techniques that can be used to study the behavior of (bio)molecules under force. Researchers in the group Sophia Gruber and Magnus Bauer developed an assay that would allow them to study the binding of the coronavirus spike protein to ACE2. They relied on two very different force spectroscopy techniques: First, they used an atomic force microscope (AFM) to pull on the proteins, a method that can apply high forces and perform fast measurements. In parallel, they employed so-called magnetic tweezers, which are extremely sensitive and can resolve very small forces. Using these methods, they imitated the forces in the mouth and throat and analyzed which forces are necessary to separate the virus from its binding partner. By using complementary force spectroscopy techniques they were able to cover the entire physiologically relevant force range.
The researchers used their new assay to compare the force stability of SARS-CoV-2 with that of SARS-CoV-1, which caused the SARS epidemic in 2002 and 2003. They found that SARS-CoV-2 could withstand significantly higher forces. “This finding explains why SARS-CoV-2 – in contrast to SARS-CoV-1 – can also affect the upper respiratory tract, where turbulent air currents generate higher forces than in the deep lungs,” says Prof. Jan Lipfert, co-author of the paper published in the journal PNAS and now professor at Utrecht University. This difference is thought to have contributed to the very different course that the 2002/03 epidemic took, compared to the current pandemic. Infections in the deep lungs with SARS-CoV-1 caused patients to become very sick more quickly, but that made it easier to isolate them and thus stop the spread of the virus.
To understand the different force stabilities in detail at the molecular level, the LMU researchers collaborated with Prof. Rafael Bernardi’s group at Auburn University in Alabama. Bernardi’s team paralleled the measured processes in so-called molecular dynamics simulations. External forces can also be applied to the spike protein in the simulations. With the help of the computer simulations, the researchers were able to understand exactly which sections of the proteins contribute to making SARS-CoV-2 more stable than SARS-CoV-1.
The LMU biophysicists were also able to show that they can use their assay to study how certain substances can prevent the coronavirus from binding to ACE2. According to the researchers, this potentially makes their measurement method a useful tool for studying neutralizing antibodies, to give just one example.
However, their main focus is currently on the new variants of the virus. Initial, as yet unpublished, results show that the different variants have different force stabilities. For example, the Alpha variant is even stronger than the original wild type of SARS-CoV-2. “It’s possible that the Omicron variant evolves in a similar way to how the original version of SARS-CoV-2 evolved compared to SARS-CoV-1. There are initial indications with Omicron that it affects the deep lung less and therefore causes less severe illness, but appears to be more transmissible.” The LMU researchers hope that their methods will be used to understand such effects in detail and even predict new variants in the future.
Magnus S. Bauer, Sophia Gruber, Adina Hausch, Priscila S.F.C. Gomes, Lukas F. Milles, Thomas Nicolaus, Leonard C. Schendel, Pilar López Navajas, Erik Procko, Daniel Lietha, Marcelo C.R. Melo, Rafael C. Bernardi, Hermann E. Gaub, and Jan Lipfert: A Tethered Ligand Assay to Probe SARS-CoV-2:ACE2 Interactions. PNAS, 2022